Please refer to Molecular Basis of Inheritance Class 12 Biology Important Questions with solutions provided below. These questions and answers have been provided for Class 12 Biology based on the latest syllabus and examination guidelines issued by CBSE, NCERT, and KVS. Students should learn these problem solutions as it will help them to gain more marks in examinations. We have provided Important Questions for Class 12 Biology for all chapters in your book. These Board exam questions have been designed by expert teachers of Standard 12.
Class 12 Biology Important Questions Molecular Basis of Inheritance
Objective Questions
Question. Which statement is true?
A. Continuous synthesis takes place on DNA strand
B. Discontinuous synthesis takes place on DNA with 5’–3’ polarity
C. In discontinuous synthesis leading strand is formed
D. In continuous synthesis lagging strand is formed
Options :
(a) A
(b) B
(c) C
(d) D
Answer
B
Question. Can only a part of DNA replicate
(a) Yes, with the help of vector in recombinant technology
(b) No
(c) Yes, with the help of RNA dependent DNA polymerase
(d) Yes, with of centrifugation technique and polymerase
Answer
A
Question. During cell cycle replication take place in
(a) G1 phase
(b) M phase
(c) G2 phase
(d) S phase
Answer
D
Question. Copying of genetic information from DNA to RNA is
(a) Replication
(b) Translation
(c) Transcription
(d) Reverse transcription
Answer
C
Question. Transcription takes place
(a) On both DNA strands simultaneously
(b) On coding strand of DNA in 5’–3’ polarity
(c) On template strand only in 5’–3’ polarity
(d) On template strand only in 3’–5’ polarity
Answer
D
Question. If a newly transcribed m RNA have a sequence 5’ AUGCAGCUC –3’ then sequence of T–RNA
(a) UACGUCAG
(b) ATGCAGTC
(c) TACGTCAG
(d) None
Answer
A
Question. According to template strand the ends if promoter and terminator
(a) 3’ end and 5’ end
(b) 5’ end and 3’end
(c) Both
(d) None
Answer
A
Question. Location of promoter is
(a) Downstream to coding strand at 5’ end
(b) Upstream to coding strand at 5’ end
(c) Downstream to coding strand at 3’ end
(d) Upstream to coding strand at 3’ end
Answer
B
Question. In process of transcription which DNA strand works as template strand
(a) Strand with 5’–3’ polarity
(a) Strand with 3’–5’ polarity
(a) Both the strands
(a) None
Answer
B
Question. Terminator sequence of transcription is located on transcription unit at :
(a) Upstream of coding strand
(b) Downstream of coding strand
(c) Upstream of template strand
(d) In between structural gene
Answer
B
Question. Meselson and Stahl separated the two DNA generated after 20 min (E.coli replicates one cycle in 20 mins) by which technique?
(a) Density gradient centrifugation
(b) X–ray diffraction
(c) Transformation
(d) None of these
Answer
A
Question. Two bands out of which one has light density and other hybrid were seen after density gradient centrifugation in Messelson and Stahl experiment were after
(a) 20 min
(b) 40 min
(c) 60 min
(d) 80 min
Answer
B
Question. Terminating factor required by RNA polymerase
(a) Rho
(b) Alpha
(c) Sigma
(d) Pi
Answer
A
Question. The replication enzyme DNA polymerase should work with
(a) High speed
(b) High accuracy
(c) None of these
(d) Both of these
Answer
D
Question. If an organism has 6 × 103 kbp (haploid content) then it will replicate in assuming average replication speed to be 2kbp/sec
(a) 45 min
(b) 500 min
(c) 50 min
(d) 100 mins
Answer
D
Very Short Answer Questions
Question. In an experiment, DNA is treated with a compound which tends to place itself amongst the stacks of nitrogenous base pairs. As a result of which the distance between two consecutive base increases, from 0.34 nm to 0.44 nm. Calculate the length of DNA double helix (which has 2 × 109 bp) in the presence of saturating amount of this compound.
Answer. 2 × 109 × 0.44 nm.
Question. Calculate the length of the DNA of bacteriophage lambda that has 48502 base pairs.
Answer. Distance between two consecutive base pairs = 0.34 × 10–9 m
The length of DNA in bacteriophage lambda = 48502 × 0.34 × 10–9 m
= 16.49 × 10–6 m
Question. How does a degenerate code differ from an unambiguous one?
Answer. Degenerate code meAnswer that one amino acid can be coded by more than one codon. Unambiguous code meAnswer that one codon codes for only one amino acid.
Question. Why is lactose considered an inducer in lac operon?
Answer. Lactose binds to repressor and prevents it from binding with the operator, as a result RNA polymerase binds to promoter–operator region to transcribe the structural genes.
Question. What is cistron?
Answer. A cistron is a segment of DNA coding for a polypeptide.
Question. State which human chromosome has
(i) the maximum number of genes and
(ii) the one which has the least number of genes?
Answer. (i) Chromosome-1
(ii) Y-Chromosome
Question. Mention two functions of the codon AUG.
Answer. Two functions of the codon AUG are:
(i) It acts as a start codon during protein synthesis.
(ii) It codes for the amino acid methionine.
Question. What is DNA fingerprinting? Mention its application.
Answer. DNA fingerprinting is the technique to determine the relationship between two DNA samples by studying the similarity and dissimilarity of VNTRs (variable number of tandem repeats). Its applications are:
(i) It is used as a tool in forensic tests to identify criminals.
(ii) To settle paternity disputes.
(iii) To identify racial groups to study biological evolution.
Question. List two essential roles of ribosome during translation.
Answer. Two essential roles of ribosome during translation are:
(i) One of the rRNA (23S in prokaryotes) acts as a peptidyl transferase ribozyme for formation of peptide bonds.
(ii) Ribosome provides sites for attachment of mRNA and charged tRNAs for polypeptide synthesis.
Question. In the medium where E. coli was growing, lactose was added, which induced the lac operon.
Then, why does lac operon shut down some time after addition of lactose in the medium?
Answer. As lac operon is an inducible operon therefore, it shuts down due to decrease in lactose substrate concentration.
Question. Explain (in one or two lines) the function of the following:
(a) Promoter (b) tRNA (c) Exons
Answer. (a) Promoter: It is the segment of DNA which lies adjacent to the operator and functions as the binding site for RNA polymerase to carry transcription if allowed by operator.
(b) tRNA: It acts as an adaptor molecule that picks up a particular amino acid from cellular pool and takes the same over to A site of mRNA for incorporation into polypeptide chain.
(c) Exons: These are the coding segments present in primary transcript which after splicing are joined to form functional mRNA.
Short Answer Questions
Question. (a) Name the enzyme responsible for the transcription of tRNA and the amino acid the initiator tRNA gets linked with.
(b) Explain the role of initiator tRNA in initiation of protein synthesis.
Answer. (a) RNA polymerase III is responsible for transcription of tRNA and the initiator tRNA gets linked with the amino acid methionine.
(b) The initiator tRNA, which is charged with amino acid methionine, reaches the smaller subunit of ribosome. Its anticodon UAC recognises the codon AUG on mRNA and binds by
forming complementary base pairs. The large subunit of ribosome joins the smaller subunit and initiates translation.
Question. Given below is a schematic representation of lac operon:
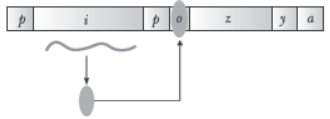
(a) Identify i and p.
(b) Name the ‘inducer’ for this operon and explain its role.
Answer. (a) i is the regulatory gene and p is the promoter gene.
(b) Lactose is the inducer. It is the substrate for the enzyme beta-galactosidase and it regulates switching on and off of the operon.
Question. How are the structural genes inactivated in lac operon in E. coli? Explain.
Answer. The regulator gene produces repressor which when free, binds to the operator region of the operon and prevents RNA polymerase from transcribing the structural genes.
Question. How are the structural genes activated in the lac operon in E. coli?
Answer. Lactose acts as the inducer that binds with repressor protein that cannot bind to operator and hence frees the operator gene. RNA polymerase freely moves over the structural genes, transcribing lac mRNA, which in turn produces the enzymes responsible for the digestion of lactose.
Question. “Genes contain the information that is required to express a particular trait.” Explain.
Answer. The genes present in an organism show a particular trait by way of forming certain product.
This is facilitated by the process of transcription and translation (according to central dogma of Biology)

Question. A low level of expression of lac operon occurs at all the time. Can you explain the logic behind this phenomena?
Answer. In the complete absence of expression of lac operon, permease will not be synthesised which is essential for transport of lactose from medium into the cells. And if lactose cannot be transported into the cell, then it cannot act as inducer. Hence, cannot relieve the lac operon from its repressed state. Therefore, lac operon is always expressed.
Question. Where is an ‘operator’ located in a prokaryote DNA? How does an operator regulate gene expression at transcriptional level in a prokaryote? Explain.
Answer. The operator region is located adjacent to promoter elements or prior to structural gene.
The operator regulates switching on and off the operon when the repressor binds to the operator region it is switched off and prevents transcription.
In the presence of inducer the repressor is inactivated and operator allows RNA polymerase to access the promoter. The operon is switched on and transcription proceeds.
Question. Would it be appropriate to use DNA probes such as VNTR in DNA fingerprinting of a bacteriophage?
Answer. Bacteriophage does not have repetitive sequence such as VNTR in its genome as its genome is very small and have all the codon sequenced. Therefore, DNA fingerprinting is not done for bacteriophages.
Question. What would happen if histones were to be mutated and made rich in amino acids aspartic acid and glutamic acid in place of basic amino acids such as lysine and arginine?
Answer. If histone proteins were rich in acidic amino acids instead of basic amino acids then they may not have any role in DNA packaging in eukaryotes as DNA is also negatively charged molecule. The packaging of DNA around the nucleosome would not happen. Consequently, the chromatin fibre would not be formed.
Question. Mention two applications of DNA polymorphism.
Answer. DNA polymorphism is applicable in genetic mapping and DNA finger printing.
Question. (a) Construct a complete transcription unit with promoter and terminator on the basis of the hypothetical template strand given below:

(b) Write the RNA strand transcribed from the above transcription unit along with its polarity.
Ans. (a)
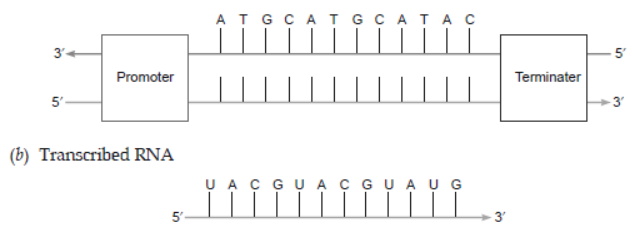
Question. (i) Name the enzyme that catalyses the transcription of hnRNA.
(ii) Why does the hnRNA need to undergo changes? List the changes that hnRNA undergoes and where in the cell such changes take place.
Answer. (i) RNA polymerase II.
(ii) hnRNA has non-functional introns in between the functional exons. To remove these, it undergoes changes. The changes that hnRNA undergoes include capping, i.e., methyl guanosine triphosphate is added to 5′ end; tailing in which poly A tail is added at 3′ end; and splicing by which introns are removed and exons are joined.
Question. One of the salient features of the genetic code is that it is nearly universal from bacteria to humAnswer. Mention two exceptions to this rule. Why are some codes said to be degenerate?
Answer. The genetic code is universal except in mitochondria and few protozo Answer.
Some codes are said to be degenerate because some amino acids are coded by more than one code.
Question. (a) Draw a clover leaf structure of tRNA showing the following:
(i) Tyrosine attached to its amino acid site.
(ii) Anticodon for this amino acid in its correct site (codon for tyrosine is UCA).
(b) What does the actual structure of tRNA look like?
Answer.

(b) The actual structure of tRNA looks like inverted L.
Long Answer Questions
Question. Draw a labelled diagram of a nucleosome. Where is it found in a cell?
Ans. Nucleosome is found in the nucleus of the cell.
Question. (a) What is this diagram representing?
(b) Name the parts a, b and c.
(c) In the eukaryotes, the DNA molecules are organised within the nucleus. How is the DNA molecule organised in a bacterial cell in absence of a nucleus?
Ans. (a) Nucleosome
(b) a—Histone octamer, b—DNA, c—H1 histone
(c) In bacterial cell, DNA in nucleoid is organised in large loops held together by proteins.
Question. (a) Draw the structure of the initiator tRNA adaptor molecule.
(b) Why is tRNA called an adaptor molecule?
Ans. (a)

(b) tRNA is called an adaptor molecule because on one end it reads the code on mRNA and on other end it would bind to the amino acid corresponding to the anticodon.
Question.
(a) Mention what enables histones to acquire a positive charge.
Ans.
(a) Basic amino acid residues of lysines and arginines.
Question. Two blood samples A and B picked up from the crime scene were handed over to the forensic department for genetic fingerprinting. Describe how the technique of genetic fingerprinting is carried out. How will it be confirmed whether the samples belonged to the same individual or to two different individuals?
Ans. Refer to Basic Concepts Point 22 (Methodology and technique).
On comparing the DNA prints of blood samples A and B, it can be confirmed that the blood sample picked up from the crime scene belongs to the same individual or to two different individuals by matching the position and thickness of the bands.
Question. Which methodology is used while sequencing the total DNA from a cell? Explain it in detail.
Ans. Methodologies of HGP:
• For sequencing, the total DNA from cell is first isolated and broken down in relatively small sizes as fragments.
• These DNA fragments are cloned in suitable host using suitable vectors. When bacteria is used as vector, they are called bacterial artificial chromosomes (BAC) and when yeast is used as vector, they are called yeast artificial chromosomes (YAC).
• Frederick Sanger developed a principle according to which the fragments of DNA are sequenced by automated DNA sequences.
• On the basis of overlapping regions on DNA fragments, these sequences are arranged accordingly.
• For alignment of these sequences, specialised computer-based programmes were developed.
• These sequences were annotated and were assigned to each chromosome. Sequence of chromosome 1 was completed only in May 2006. It was the last chromosome be sequenced).
• Finally, the genetic and physical maps of the genome were constructed by collecting information about certain repetitive DNA sequences and DNA polymorphism, based on endonuclease recognition sites.
Question. List the salient features of double helix structure of DNA.
Ans. Following are some features of DNA:
(i) DNA is made up of two polynucleotide chains, where the backbone is made up of sugar and phosphate groups and the nitrogenous bases project towards the centre.
(ii) There is complementary base pairing between the two strands of DNA. ‘A’ pairs with ‘T’ with 2 hydrogen bonds and ‘C’ pairs with ‘G’ with 3 hydrogen bonds.
(iii) The two strands are coiled in right-handed fashion and are anti-parallel in orientation. One chain has a 5′→3′ polarity while the other has 3′→5′ polarity.
(iv) The diameter of the strand is always constant due to pairing of purine and pyrimidine, i.e.,adenine is complementary to thymine while guanine is complementary to cytosine.
(v) The distance between two base pairs in a helix is 0.34 nm and a complete turn contains approximately ten base pairs. The pitch of the helix is 3.4 nm and the two strands are righthanded coiled.
(vi) Plane of one base pair stacks over the other in double helix. This in addition to hydrogen bonds confers stability to the helical structure.
Question. It is established that RNA is the first genetic material. Explain giving three reasons.
Ans. The reasons in support are:
(i) Processes like metabolism, translation and splicing evolve around RNA.
(ii) RNA is reactive and catalyses reactions.
(iii) In some viruses, RNA is the hereditary material.
(iv) RNA is unstable and can be easily mutated leading to evolution. (Any three)
Question. A typical mammalian cell has 22 metres long DNA molecule whereas the nucleus in which it is packed measures about 10–6 m. Explain how such a long DNA molecule is packed within a tiny nucleus in the cell.
Ans. Packaging of DNA in eukaryotes
• Roger Kornberg (1974) reported that chromosome is made up of DNA and protein.
• Later, Beadle and Tatum reported that chromatin fibres look like beads on the string, where beads are repeated units of proteins.
• The proteins associated with DNA are of two types— basic proteins (histones) and acidic non-histone chromosomal (NHC) proteins.
• The negatively charged DNA molecule wraps around the positively charged histone proteins to form a structure called nucleosome.
• The nucleosome core is made up of four types of histone proteins—H2A, H2B, H3 and H4 occurring in pairs..
Question. Answer the following questions based on Meselson and Stahl’s experiment:
(a) Write the name of the chemical substance used as a source of nitrogen in the experiment by them.
(b) Why did the scientists synthesise the light and the heavy DNA molecules in the organism used in the experiment?
(c) How did the scientists make it possible to distinguish the heavy DNA molecule from the light DNA molecule? Explain.
(d) Write the conclusion the scientists arrived at after completing the experiment.
Ans. (a) Ammonium chloride (NH4Cl).
(b) To check if DNA replication was semi-conservative.
(c) The heavy and light DNA molecules were distinguished by centrifugation in a caesium chloride density gradient.
(d) The scientists concluded that DNA replicates semi-conservatively.
Question. How are the DNA fragments separated and isolated for DNA fingerprinting? Explain.
Ans. Separation and Isolation of DNA Fragments (Gel Electrophoresis)
• Gel electrophoresis is a technique for separating DNA fragments based on their size.
• Firstly, the sample DNA is cut into fragments by restriction endonucleases.
• The DNA fragments being negatively charged can be separated by forcing them to move towards the anode under an electric field through a medium/matrix.
• Commonly used matrix is agarose, which is a natural linear polymer of D-galactose and 3, 6-anhydro-L-galactose which is extracted from sea weeds.
• The DNA fragments separate-out (resolve) according to their size because of the sieving property of agarose gel. Hence, the smaller the fragment size, the farther it will move.
• The separated DNA fragments are visualised after staining the DNA with ethidium bromide followed by exposure to UV radiation.
• The DNA fragments are seen as orange coloured bands.
• The separated bands of DNA are cut out and extracted from the gel piece. This step is called elution.
• The purified DNA fragments are used to form recombinant DNA which can be joined with cloning vectors.
Question. The base sequence in one of the strands of DNA is TAGCATGAT.
(i) Give the base sequence of its complementary strand.
(ii) How are these base pairs held together in a DNA molecule?
(iii) Explain the base complementarity rules. Name the scientist who framed this rule.
Ans. (i) The complementary strand is ATCGTACTA.
(ii) The base pairs are held together by hydrogen bonds in a DNA molecule. A and T are held by two hydrogen bonds while G and C are held by three hydrogen bonds.
(iii) Watson and Crick framed the base complementarity rule. The rule states that the ratios between adenine and thymine, and guanine and cytosine are constant and equals one.
Question. Describe the experiments that established the identity of ‘transforming principles’ of Griffith.
Ans. Oswald Avery, Colin MacLeod and Maclyn McCarty purified biochemicals (proteins, DNA, RNA, etc.) from the heat-killed S cells to see which ones could transfrom live R cells into S cells.
They discovered that DNA alone from S bacteria caused R bacteria to become transformed.
They also discovered that protein-digesting enzymes (proteases) and RNA-digesting enzymes (RNases) did not affect transformation, so the transforming substance was not a protein or RNA. Digestion with DNase did inhibit transformation, suggesting that the DNA caused the transformation. They concluded that DNA is the hereditary material.
Question. Describe the experiment that proved that DNA is the genetic material.
Ans. • Procedure:
(i) Some bacteriophage virus were grown on a medium that contained radioactive phosphorus (32P) and some in another medium with radioactive sulphur (35S).
(ii) Viruses grown in the presence of radioactive phosphorus (32P) contained radioactive DNA.
(iii) Similar viruses grown in presence of radioactive sulphur (35S) contained radioactive protein.
(iv) Both the radioactive virus types were allowed to infect E. coli separately.
(v) Soon after infection, the bacterial cells were gently agitated in blender to remove viral coats from the bacteria.
(vi) The culture was also centrifuged to separate the viral particle from the bacterial cell.
• Observations and Conclusions:
(i) Only radioactive 32P was found to be associated with the bacterial cell, whereas radioactive 35S was only found in surrounding medium and not in the bacterial cell.
(ii) This indicates that only DNA and not the protein coat entered the bacterial cell.
(iii) This proves that DNA is the genetic material which is passed from virus to bacteria and not protein.
Question. In a series of experiments with Streptococcus and mice, F. Griffith concluded that R-strain bacteria had been transformed. Explain.
Ans. • Frederick Griffith (1928) conducted experiments with Streptococcus pneumoniae (bacterium causing pneumonia).
• He observed two strains of this bacterium—one forming smooth shiny colonies (S-type) with capsule, while other forming rough colonies (R-type) without capsule.
Question. How was a heavy isotope of nitrogen used to provide experimental evidence to semiconservative mode of DNA replication?
OR
Describe the experiment that helped to demonstrate the semi-conservative mode of DNA replication.
Ans.

Question. With respect to Messelson and Stahl’s Experiment, answer the following questions:
(a) Identify the method used to distinguish between heavy and light isotopes of nitrogen.
(b) With the help of diagrams, compare the results for the DNA isolated after 20 minutes of experiment with the DNA which was isolated after 40 minutes.
Ans. (a) Centrifugation in a CsCl density gradient.
(b)

Question. What are the three types of RNA? Mention their relation to protein synthesis.
Ans. Types of RNA
| S.No. | Types of RNA | Functions |
| (i) | Messenger RNA (mRNA) | (i) It stores the genetic information from DNA. (ii) It decides the sequence of amino acid in a polypeptide. |
| (ii) | Transfer RNA (tRNA) | (i) It acts as an adaptor molecule that at one end reads the code on mRNA and accordingly bind to amino acid on the other end. (ii) It recognises the codon on mRNA by its anticodon and leaves amino acid at the protein synthesis site. |
| (iii) | Ribosomal RNA (rRNA) | (i) It constitutes the ribosomal structure.(ii) It helps to form peptide bond. |
Question. (a) Differentiate between a template strand and coding strand of DNA.
(b) Name the source of energy for the replication of DNA.
Ans.
(b) Deoxynucleoside triphosphates provide the energy for DNA replication.
Question. Draw a labelled schematic sketch of replication fork of DNA. Explain the role of the enzymes involved in DNA replication.
Ans. Enzymes involved in DNA replication are:
(i) DNA-dependent DNA polymerase, which catalyses the polymerisation of polynucleotides in a very short time only in 5′→3′ direction with accuracy.
(ii) DNA ligase, which joins the discontinuously synthesised short segments called Okazaki fragments formed on one of the template strands.
Question. (a) State a reason why is the replication continuous and discontinuous in the diagram drawn.
Ans.
(a) The two strands of DNA are anti parallel, i.e., one strand runs in the direction 5′ to 3′ and the other runs in the direction 3′ to 5′. DNA polymerase adds deoxyribonucleotides only in one direction, i.e., 5′ to 3′. Thus, producing Okazaki fragments in the other strands.
Question. (a) What is an operon?
(b) Explain how a polycistronic structural gene is regulated by a common promoter and a combination of regulatory genes in a lac operon.
Ans. (a) An operon is a polycistronic structural gene which is regulated by a common promoter and regulator gene.
(b) • Operon: The concept of operon was first proposed in 1961, by Jacob and Monod. An operon is a unit of prokaryotic gene expression which includes coordinately regulated (structural) genes and control elements which are recognised by regulatory gene product.
• The lactose operon: The lac z, lac y, lac a genes are transcribed from a lac transcription unit under the control of a single promoter. They encode enzyme required for the use of lactose as a carbon source.The lac i gene product, the lac repressor, is expressed from a separate transcription unit upstream from the operator.
• Regulation of lac operon by repressor is referred to as negative regulation.
• lac operon consists of three structural genes (z, y, a), operator (o), promoter (p) and a separate regulatory gene (i). Lactose is the inducer in lac operon.
Question. Study the schematic representation of the genes involved in the lac operon given below and answer the questions that follow:

(i) Identify and name the regulatory gene in this operon. Explain its role in ‘switching off’ the operon.
(ii) Why is lac operon’s regulation referred to as negative regulation?
(iii) Name the inducer molecule and the products of the genes ‘z’ and ‘y’ of the operon. Write the functions of these gene products.
Ans. (i) i gene is the regulatory gene and codes of repressor which acts as inhibitor as inhibits the transcription of structural genes.
The repressor of the operon is synthesised from the i gene. The repressor protein in the absence of an inducer (lactose or allolactose) binds to the operator region of the operon and
prevents RNA polymerase from transcribing the structural genes. Thus ‘switching off’ the operon.
(ii) Regulation by lac operon is referred to as negative regulation because the repressor binds to the operator for ‘switching off’ the operon.
(iii) Lactose or allolactose acts as an inducer. Gene z codes for b-galactosidase (gal) enzyme which breaks lactose into galactose and glucose. Gene y codes for permease, which increases the permeability of the cell to lactose.
Question. (a) Absence of lactose in the culture medium affects the expression of a lac operon in E. coli. Why and how? Explain.
(b) Write any two ways in which the gene expression is regulated in eukaryotes.
Ans. (a) (i) When lactose is absent, i gene regulates and produces repressor mRNA which translate repression.
(ii) The repressor protein binds to the operator region of the operon and as a result prevents RNA polymerase to bind to the operon.
(iii) The operon is switched off.
(b) Gene expression in eukaryotes is regulated at following levels:
Transcriptional level (formation of primary transcripts)
Processing level (regulation of splicing)
Transport of messenger RNA from nucleus to the cytoplasm
Translational level. (Any two)
Question. (i) DNA polymorphism is the basis of DNA fingerprinting technique. Explain.
(ii) Mention the causes of DNA polymorphism.
Ans. (i) Allelic sequence variation has traditionally been described as a DNA polymorphism if its frequency is greater than 0.01. Simply, if an inheritable mutation is observed in a population at high frequency, it is referred to as DNA polymorphism. DNA fingerprinting is a technique of determining nucleotide sequences of certain areas of DNA which are unique to each individual.
Although the DNA from different individuals is more alike than different, there are many regions of the human chromosomes that exhibit a great deal of diversity. Such variable sequences are termed “polymorphic” (meaning many forms) A special type of polymorphism, called VNTR (variable number of tandem repeats), is composed of repeated copies of a DNA sequence that lie adjacent to one another on the chromosome. Since polymorphism is the basis of genetic mapping of human genome, therefore it forms the basis of DNA fingerprinting too.
(ii) The probability of such variations to be observed in non-coding DNA sequences would be higher as mutations in these sequences may not have any immediate effect in an individual’s reproductive ability. These mutations keep on accumulating generation after generation and form one of the basis of variability. There is a variety of different types of polymorphisms ranging from single nucleotide change to very large scale changes. For evolution and speciation, such polymorphisms play very important role.
The single nucleotide polymorphisms are used in locating diseases and tracing of human history.
DNA polymorphisms are due to mutations.